
HPV Detection in Leukocyte Samples of Spinocellular Carcinomas Using PCR
Ana Maria Jimenez Jimenez, Lourdes Maria Herrera-Quintana, Simona Dostalova, Zbynek Heger, Branislav Ruttkay-Nedecky, and Rene Kizek
Head and neck squamous cell carcinomas (HNSCCs) are malignant tumors of epidermal keratinocytes, that affect mainly the lip, oral cavity, nasal cavity, paranasal sinuses, pharynx, and larynx [1]. HNSCCs grow locally and infrequently metastasize [2]. This carcinomas are one of the major forms of skin cancer, which arises from the uncontrolled multiplication of epithelial cells or other cell types such as keratinocytes, tonofilament bundles, desmosomes, or structures involved in cell-to-cell adhesion [3]. At the time of diagnosis, HNSCCs spread to the lymph nodes of the neck, and this is often the first sign of the disease. The surgery and radiation are the common therapy against this cancer [4].
The HNSCCs is the sixth leading cancer by incidence worldwide and eighth by death. There are 0.5 million new cases a year worldwide [5]. The environmental pollution and wrong life style, which are associated with the squamous cell carcinomas, involve following risk factors: tobacco smoking, alcohol consumption, UV light, particular chemicals used in certain workplaces, and certain strains of viruses, such as human papillomavirus (HPV) [6].
Human papillomaviruses (HPVs) are small DNA viruses with a strict tropism for human epithelial cells. In general, HPV infections are asymptomatic and persist for 18–24 months before they are cleared by the immune system of the host [7]. Tumorigenicity of HPVs differs markedly among HPV genotypes [8]. This virus has been associated with HNSCCs in the areas of the oropharynx, lung, fingers, and anogenital region [1]. Functional loss of p53 tumor suppressor gene, mutations in Ras protooncogenes, and certain chromosomal aberrations are characteristics of cancer caused by HPVs [9].
Many different methods are able to give information about the presence of HPVs in biological samples. The combination of a sensitive test, and a specific test, could allow for the best potential to accurately establish the presence or absence of HPVs [10,11].The mainly utilized tests for HPV identification among others, are polymerase chain reaction (PCR) testing, real time PCR, in situ hybridization analysis (IS), immunohistochemical (IHC) staining for tumor suppressor protein p16, and southern blotting assays [11].
The methodology for polymerase chain reaction (PCR) is an indispensable technique used in molecular biology in research labs to amplify a single copy of DNA generating thousands to millions of copies of chosen DNA sequence [12]. This technique has a big variety of applications like DNA cloning for DNA sequencing, DNA-based phylogeny, or functional analysis of genes; the diagnosis of hereditary diseases; the identification of genetic fingerprints and the detection and diagnosis of infectious diseases. PCR can be extensively modified to perform a wide array of genetic manipulations [10,13].
In HPV detection the PCR represents a highly-sensitive and cost-effective method. It can be used to detect as little as one copy of a DNA sequence from paraffin-embedded tissues or fresh tissues from biopsies [14]. The disadvantages of PCR techniques are that they have lower specificity, they do not allow distinction between all HPV types that are present in the neoplastic cells and non-neoplastic and they cannot distinguish between episomal and integrated HPV DNA[15]. Furthermore, the presence of latent viruses leads to false positive results due to the ability of PCR to detect just a few copies of HPV DNA per cell. Different attempts have been made to resolve this issue through the use of real-time PCR, which provides a quantitative analysis of viral load [16].
The combined results of immunocytochemical detection of p16 protein and the detection of HPV DNA by specific PCR enable better discrimination between latent and carcinogenic HPV infections and thus can provide information on the prognosis of HNSCC patients and facilitate therapeutic decisions[17].
The detection of the viral oncoproteins E6 and E7 requires technique that is restricted to the research laboratory, like RNA extraction and polymerase chain reaction amplification. The development of RNA in situ hybridization (ISH) probes complementary to E6/E7 mRNA permits direct visualization of viral transcripts in routinely processed tissues and has opened the door for accurate HNSCC detection in the clinical care setting [18].
![Figure 1: Lecture by Associate Professor Vojtech
Adam of Mendel University in Brno on the role of
metallothionein in cancer [1]. houbova](pics/jimenez1.jpg)
Table 1: Clinical characterization of patients. Analysis the medical information obtained from patients with head and neck cancer from St. Anne´s University Hospital in Brno, differentiating between men and women and their clinical histological data.
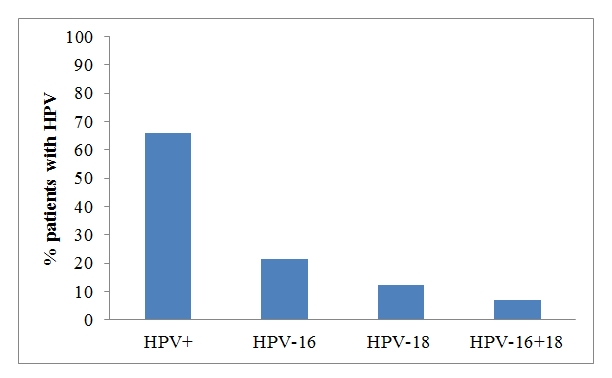
Figure 2: HPV detection and subtypes identification of the patients by PCR method. GP5/GP6 primers were used for HPV detection (65.9%) and HPV16-18 primer for subtyping (21.4% HPV-16, 12.5% HPV-18 and a 7% for HPV16-18).
![Figure 1: Lecture by Associate Professor Vojtech
Adam of Mendel University in Brno on the role of
metallothionein in cancer [1]. houbova](pics/jimeneze2.jpg)
Figure 1: Scheme of HPV detection using PCR. First, GP5/GP6 primers for HPV detection were used. Next, the 56 HPV positive samples were analyzed with the type-specific primers for HPV-16 and HPV-18, obtaining 12 samples of HPV16, 7 samples of HPV-18 and 4 samples were HPV-16 and HPV-18 at the same time.

Figure 3: Multiple sequence alignment using ClustalW2. The sequences used in this alignment were, sequence of the amplicon obtained with the GP5/GP6 set of primers (called database), the sequence obtained from analysis of our samples (called Sequence), and a complete sequence of L1 gene (called L1 complete).
![Figure 1: Lecture by Associate Professor Vojtech
Adam of Mendel University in Brno on the role of
metallothionein in cancer [1]. houbova](pics/jimeneze5.jpg)
Table 2: List of primers used in this study [25].
1. Syrjanen, S. Human papillomavirus (hpv) in head and neck cancer. J. Clin. Virol. 2005, 32, S59-S66.
2. Syrjanen, S.; Rautava, J. Hpv and oral health response. J. Am. Dent. Assoc. 2012, 143, 442-444.
3. Syrjanen, K.J.; Chang, F.; Syrjanen, S.M. Hpv infections in etiology of benign and malignant sinonasal, bronchial and oesophageal squamous cell lesions. Medimond S R L: 40128 Bologna, 2000; p 169-179.
4. Janicek, M.F.; Averette, H.E. Cervical cancer: Prevention, diagnosis, and therapeutics. CA-Cancer J. Clin. 2001, 51, 92-114.
5. Badulescu, F.; Crisan, A.; Badulescu, A.; Schenker, M. Recent data about the role of human papillomavirus (hpv) in oncogenesis of head and neck cancer. Rom. J. Morphol. Embryol. 2010, 51, 437-440.
6. Chen, A.Y.; DeSantis, C.; Jemal, A. Us mortality rates for oral cavity and pharyngeal cancer by educational attainment. Arch. Otolaryngol. Head Neck Surg. 2011, 137, 1094-1099.
7. Schwartz, S. Papillomavirus transcripts and posttranscriptional regulation. Virology 2013, 445, 187-196.
8. Tsao, K.-C.; Huang, C.-G.; Kuo, Y.-B.; Chang, T.-C.; Sun, C.-F.; Chang, C.A.; Yang, S.-L.; Chan, E.-C. Prevalence of human papillomavirus genotypes in northern taiwanese women. Journal of Medical Virology 2010, 82, 1739-1745. 9. Garnett, T.O.; Duerksen-Hughes, P.J. Modulation of apoptosis by human papillomavirus (hpv) oncoproteins. Arch. Virol. 2006, 151, 2321-2335.
10. Gagnon, D.; Fradet-Turcotte, A.; Archambault, J. A quantitative and high-throughput assay of human papillomavirus DNA replication. Methods in molecular biology (Clifton, N.J.) 2015, 1249, 305-316.
11. Venuti, A.; Paolini, F. Hpv detection methods in head and neck cancer. Head and neck pathology 2012, 6 Suppl 1, S63-74.
12. Haugg, A.M.; Rennspiess, D.; zur Hausen, A.; Speel, E.J.M.; Cathomas, G.; Becker, J.C.; Schrama, D. Fluorescence in situ hybridization and qpcr to detect merkel cell polyomavirus physical status and load in merkel cell carcinomas. Int. J. Cancer 2014, 135, 2804-2815.
13. Rodel, F.; Wieland, U.; Fraunholz, I.; Kitz, J.; Rave-Frank, M.; Wolff, H.A.; Weiss, C.; Wirtz, R.; Balermpas, P.; Fokas, E., et al. Human papillomavirus DNA load and p16(ink4a) expression predict for local control in patients with anal squamous cell carcinoma treated with chemoradiotherapy. International journal of cancer. Journal international du cancer 2015, 136, 278-288.
14. Heidegger, I.; Pichler, R.; Muller, B.; Klocker, H.; Oswald, D.; Haid, B.; Zelger, B.; Horninger, W.; Oswald, J. Is real-time pcr the correct method to evaluate the incidence of human papillomavirus in prepuces of asymptomatic boys and men? World J. Urol. 2014, 32, 1199-1204.
15. Micalessi, M.I.; Boulet, G.A.; Bogers, J. A real-time pcr approach based on spf10 primers and the inno-lipa hpv genotyping extra assay for the detection and typing of human papillomavirus. Methods in molecular biology (Clifton, N.J.) 2015, 1249, 27-35.
16. Sahiner, F.; Kubar, A.; Gumral, R.; Ardic, M.; Yigit, N.; Sener, K.; Dede, M.; Yapar, M. Efficiency of my09/11 consensus pcr in the detection of multiple hpv infections. Diagn. Microbiol. Infect. Dis. 2014, 80, 43-49.
17. Linxweiler, M.; Bochen, F.; Wemmert, S.; Lerner, C.; Hasenfus, A.; Bohle, R.M.; Al-Kadah, B.; Takacs, Z.F.; Smola, S.; Schick, B. Combination of p16(ink4a)/ki67 immunocytology and hpv polymerase chain reaction for the noninvasive analysis of hpv involvement in head and neck cancer. Cancer Cytopathol. 2015, 123, 219-229.
18. Westra, W.H. Detection of human papillomavirus (hpv) in clinical samples: Evolving methods and strategies for the accurate determination of hpv status of head and neck carcinomas. Oral Oncol. 2014, 50, 771-779.
19. Jacobs, M.V.; Snijders, P.J.F.; vandenBrule, A.J.C.; Helmerhorst, T.J.M.; Meijer, C.; Walboomers, J.M.M. A general primer gp5+/gp6+-mediated pcr-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. Journal of Clinical Microbiology 1997, 35, 791-795.
20. De Roda Husman, A.-M.; Walboomers, J.M.M.; Van Den Brule, A.J.C.; Meijer, C.J.L.M.; Snijders, P.J.F. The use of general primers gp5 and gp6 elongated at their 3‘ ends with adjacent highly conserved sequences improves human papillomavirus detection by pcr. J. Gen. Virol. 1995, 76, 1057-1062.
21. Baay, M.F.D.; Quint, W.G.V.; Koudstaal, J.; Hollema, H.; Duk, J.M.; Burger, M.P.M.; Stolz, E.; Herbrink, P. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by pcr in paraffin-embedded cervical carcinomas. Journal of Clinical Microbiology 1996, 34, 745-747.
22. Snijders, P.J.F.; Vandenbrule, A.J.C.; Schrijnemakers, H.F.J.; Snow, G.; Meijer, C.; Walboomers, J.M.M. The use of general primers in the polymerase chain-reaction permits the detection of a broad-spectrum of human papillomavirus genotypes. J. Gen. Virol. 1990, 71, 173-181.
23. Soler, C.; Allibert, P.; Chardonnet, Y.; Cros, P.; Mandrand, B.; Thivolet, J. Detection of human papillomavirus type-6, type-11, type-16 and type-18 in mucosal and cutaneous lesions by the multiplex polymerase chain-reaction. J. Virol. Methods 1991, 35, 143-157.
24. Ueyama, H.; Kurokawa, K.; Sasaki, I.; Ueda, K. Characterization of acidic actin in mouse sarcoma-180 cells. Cell Structure and Function 1987, 12, 463-470.
25. Husnjak, K.; Grce, M.; Magdic, L.; Pavelic, K. Comparison of five different polymerase chain reaction methods for detection of human papillomavirus in cervical cell specimens. J. Virol. Methods 2000, 88, 125-134.
26. Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389-3402.
27. Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R., et al. Clustal w and clustal x version 2.0. Bioinformatics 2007, 23, 2947-2948.
J.Met.Nano:
volume-2, issue-3
- Laboratory of Metallomics and Nanotechnologies – an initiator of the Metallomics Scientific Network formation
- Capillary electrophoresis of metallothionein
- Synthetic birnessites and buserites as heavy metal cation traps and environmental remedies
- Immunohistochemical detection of metallothionein
- MALDI-TOF MSI and electrochemical detection of metallothionein in chicken liver after cadmium exposure
- The use of MALDI MSI for the study of different tissues
- Utilization of graphene oxide electrophoretic deposition for construction of electrochemical sensors and biosensors
- Influence of Different Inducers on Ligninolytic Enzyme Activities
- Interaction of nanocarrier apoferritin with cytotoxic drug molecules
- Study of cell penetrating peptide and Europium(III) and Terbium(III) Schiff base complexes interaction
- HPV Detection in Leukocyte Samples of Spinocellular Carcinomas Using PCR
- Characterization of carbon quantum dots by capillary electrophoresis with laser-induced fluorescence detections
- Modification of anti-DNA antibodies with carbon quantum dots
- Fluorescence detection of carbon quantum dots assessed by stratospheric platform
 PDF
PDF